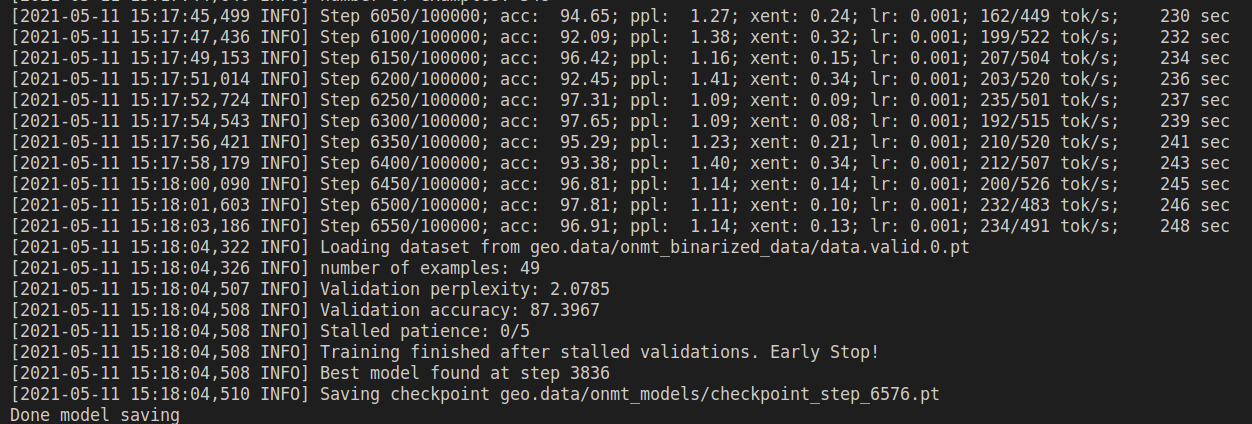
Originally I tried the seq2seq model (Glove embedding + RNN encoder-decoder + copy generator) on Text2SQL task by OpenNMT, everything works perfectly fine. I can get an accuracy of ~60% on the GeoQuery benchmark, the cross-entropy on the training set will drop to as low as 0.10, and accuracy on training will be something > 90% (token level accuracy).
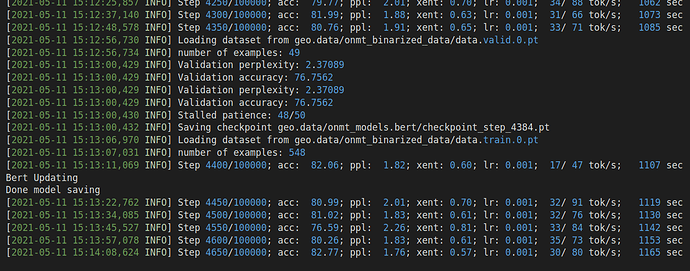
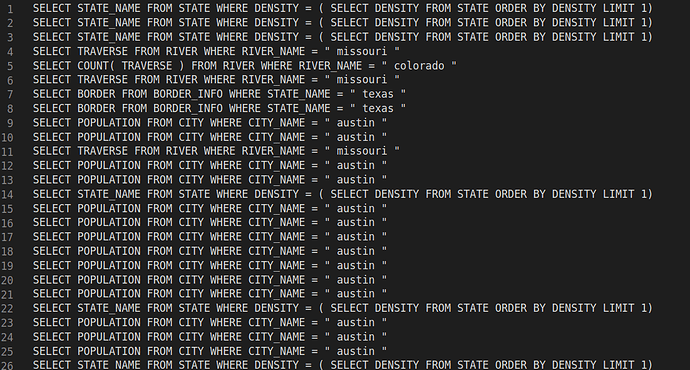
When I add Bert encoder and replace the Glove embedding with the last layer output of Bert on the encoder, the model seems to learn nothing during training. The token level accuracy in training cannot reach 90%, and the cross-entropy will remain something like 0.3. During inference, the model predicts unreasonable SQL results and can barely achieve 1% accuracy on the testing set.
I have investigated this issue for quite a long time, I double-checked my optimizer, and I use different optimizer (Adam with a learning rate of 1e-3 for parameters in my LSTM part, BertAdam with a learning rate of 1e-5 for Bert part). For the encoding part, I directly copy codes from a published Github Repo.
I could not come up with any other places that my code might go wrong. Any help will be much appreciated!
BTW, I have posted my training information as well as the SQL prediction results in the reply session for your reference.