hello
I use fast_ Align generates alignment files.
Pretreatment increase --train_align --valid_align
Complete command:
onmt_preprocess -src_seq_length 260 -tgt_seq_length 260 -filter_valid -train_align train_align.txt -valid_align valid_align.txt -train_src tgt-train.txt -train_tgt src-train.txt -tgt-val.txt -valid_tgt src-val.txt -save_data pretreatment/demo -overwrite
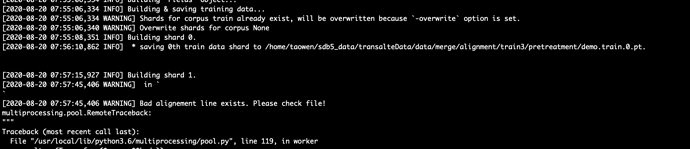
But get wrong:
I don’t know where the problem is. Check that there are no blank lines in the file.
Asking for help.